La Biologie des Systèmes dans le Carcinome Rénal
(Group : Regulatory Networks of angiogenesis, Tumor Invasion and Metastasis)
Renal Cell Carcinoma (RCC)
Renal Cell Carcinoma (RCC) accounts for 2-3% of adult malignancies and has a world-wide mortality rate of over 100,000 people per year. Incidence and prevalence are rising along with related risk factors such as obesity, diabetes and hypertension. RCC is resistant to standard chemotherapeutic agents and radiotherapy, Standard treatment is surgery plus antiangiogenic agents, but therapy response is variable, and with no biomarkers validated for clinical use, accurate prediction of disease-free survival is difficult. RCC metastasises primarily to the lung (secondarily to liver and bone), and five year survival in the presence of metastases is less than 10%5. Immunotherapy confers a small but significant survival advantage in metastatic RCC, but is effective in only a small minority of patients4. The pathophysiology of RCC is still far from understood, and there is a clear need to identify key mechanisms of tumour progression and metastasis to open up novel therapeutic avenues targeting different aspects of RCC biology. Our ability to identify novel molecular mechanisms underlying the development of primary and metastatic tumours is still unsatisfactory. It has been shown that Systems Biology approaches designed for the analysis of multi-level functional genomics datasets can be very successful, and we hypothesise that these approaches will allow us to identify and exploit gene networks involved in RCC pathogenesis.
The model
Using syngeneic mouse models of renal carcinoma combined with in-depth bioinformatics analysis and computational modelling of genomic and expression data, we are generating high resolution data sets to couple molecular changes with mechanisms underlying renal carcinoma development and metastasis. Candidate genes and genetic signatures identified and tested using this approach are then tested for relevance in human patient samples in concert with our clinical team. We already have one target gene which has good potential as a prognostic marker and putative therapeutic target. The final aim of the project is to integrate the data obtained from molecular network discovery, key gene functional analyses, patient data and tumour imaging into mathematical models capable of representing the processes of tumour formation and metastasis, and predicting patient outcome.
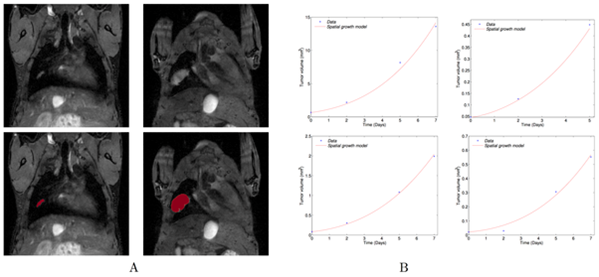 |
| We derived a mathematical model of spatial tumour growth, confronted it with experimental data of single metastatic tumour growth, and used it to provide insights on the dynamics of multiple tumours growing in close vicinity. Together, our results have implications for theories of the metastatic process and suggest that global dynamics of metastasis development is dependent on spatial interactions between metastatic lesions (Baratchat et al, 2015). |
Project members :
– Prof. Andreas Bikfalvi (Project Investigator)
– Dr. Lin Cooley
– Dr. Wilfried Souleyreau
Collaborations :
– F. Falciani, Institute of Integrative Biology, University Liverpool.
– T. Colin, MONC, Mathematics Institute, Bordeaux.
Publications associated to the project :
Computational Modelling of Metastasis Development in Renal Cell Carcinoma. Baratchart E, Benzekry S, Bikfalvi A, Colin T, Cooley LS, Pineau R, Ribot EJ, Saut O, Souleyreau W. PLoS Comput Biol. 2015 Nov 23;11(11):e1004626. doi: 10.1371/journal.pcbi.1004626. eCollection 2015 Nov. PMID: 26599078

